In the first part of this project, the Bill and Melinda Gates Foundation (BMGF) and the Burden of Disease Working Group of the World Health Organization (WHO) commissioned the Strategic Analysis, Research, and Training Center (START) to perform a systematic review of literature to better understand the relationship between the presence of pathogens and the occurrence of diarrhea. START generated a dataset of 170 case-control and cohort studies of diarrhea-associated enteric pathogens published between 1990 and 2019 that present pathogen specific risk estimates.
The studies encompass data for 15 enteric pathogens and included more than 60,000 cases and controls from case-control studies and 60,000 diarrheal samples and 200,000 non-diarrheal samples from cohort studies. The most commonly detected pathogens in the included studies were Campylobacter, ETEC, rotavirus, and Shigella. The dataset can be used to generate aggregate measures of association stratified by age group, year of data collection, laboratory detection method, country rotavirus immunization status, clinical severity, strain of pathogen, and community or health facility setting. START anticipates that this dataset will be valuable to the WHO Burden of Disease working group in identifying heterogeneity in the risk of diarrhea from specific pathogens in different populations.
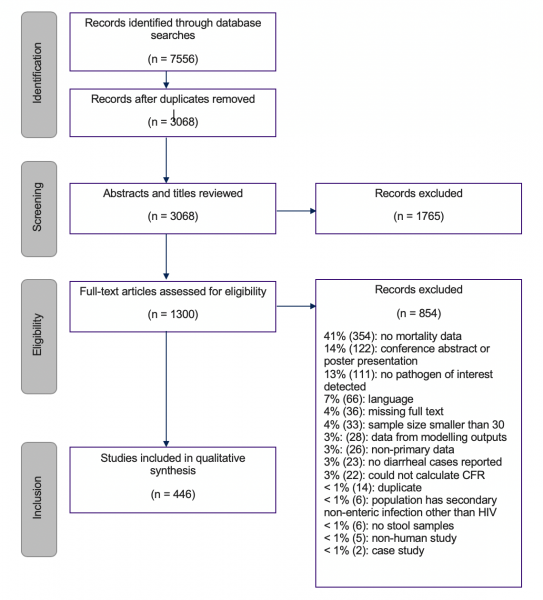 The second part of this project conducted a systematic review of publications to estimate case fatality risks (CFRs) of specific pathogens in various settings in order to understand the level of heterogeneity of CFRs across pathogen, countries, regions, and treatment settings. The review was to pay particular attention to both hospital and community data to capture potential bias in previous estimates, as well as to gather data on sampling time frames, time to death, treatments given, and detection methods. Studies were selected for inclusion if they presented primary data, if their participants had diarrhea and an identified pathogen of interest, and if they either included information about the number of deaths or presented follow-up data for all participants, allowing the team to deduce that no participants died due to enteric pathogens during the study. Two independent reviewers screened each title and abstract, and then later the full text of each study. The variables extracted included:
The second part of this project conducted a systematic review of publications to estimate case fatality risks (CFRs) of specific pathogens in various settings in order to understand the level of heterogeneity of CFRs across pathogen, countries, regions, and treatment settings. The review was to pay particular attention to both hospital and community data to capture potential bias in previous estimates, as well as to gather data on sampling time frames, time to death, treatments given, and detection methods. Studies were selected for inclusion if they presented primary data, if their participants had diarrhea and an identified pathogen of interest, and if they either included information about the number of deaths or presented follow-up data for all participants, allowing the team to deduce that no participants died due to enteric pathogens during the study. Two independent reviewers screened each title and abstract, and then later the full text of each study. The variables extracted included:
-
- Basic publication information
- Quality indicators
- Pathogen(s) detected, including coinfections within a single patient
- Pathogen detection method
- Age of participants
- Study setting
- Country rotavirus vaccination status
- Severity
- Participant HIV status
- Treatment given
- Presentation of diarrhea
- Cases
- Deaths attributable to diarrhea as indicated by the authors
- Time relative to hospitalization when fecal sample was taken
- Follow-up time
- Time to death
Because of the primary interest in pathogen specificity, the data were extracted such that pathogen-specific CFRs can be aggregated.